Which Of The Following Images Represents Newly Synthesized Dna That Can Be Repaired
Let us make an in-depth study of the DNA damage types and repair mechanisms. The damage types of DNA are: one. Simple Mutations 2. Deamination 3. Missing Bases 4. Chemical Modification of Bases 5. Formation of Pyrimidine Dimers (Thymine Dimers) and 6. Strand Breaks. The various DNA repair mechanisms are: one. Direct Repair ii. Excision Repair iii. Mismatch Base Repair 4. Recombination Repair or Retrieval Organisation and five. SOS Repair Mechanism.
Introduction to DNA Impairment and Repair:
DNA is a highly stable and versatile molecule. Though sometimes the damage is caused to it, it is able to maintain the integrity of data independent in it. The perpetuation of genetic cloth from generation to generation depends upon keeping the rates of mutation at low level. DNA has many elaborate mechanisms to repair any impairment or distortion.
The nearly frequent sources of harm to Dna are the inaccuracy in DNA replication and chemical changes in DNA. Malfunction of the process of replication can lead to incorporation of wrong bases, which are mismatched with the complementary strand. The damage causing chemicals pause the backbone of the strand and chemically modify the bases. Alkylation, oxidation and methylation cause damage to bases. 10-rays and gamma radiations crusade unmarried or double stranded breaks in Deoxyribonucleic acid.
A change in the sequence of bases if replicated and passed on to the next generation becomes permanent and leads to mutation. At the same time mutations are too necessary which provide raw material for evolution. Without evolution, the new species, fifty-fifty man beings would not have arisen. Therefore a balance between mutation and repair is necessary.
Types of Impairment:
Damage to DNA includes any deviation from the usual double helix construction.
one. Simple Mutations:
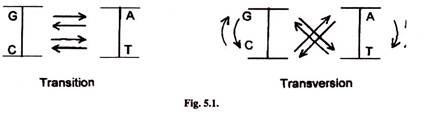
Simplest mutations are switching of one base for another base of operations. In transition one pyrimidine is substituted by another pyramidine and purine with some other purine. Trans-version involves substitution of a pyramidine by a purine and purine by a pyrimidine such every bit T past G or A and A past C or T. Other simple mutations are detection, insertion of a single nucleotide or a small number of nucleotides. Mutations which modify a single nucleotide are called point mutations.
2. Deamination:
The common alteration of course or impairment includes deamination of cytosine (C) to course uracil (u) which base pairs with adenine (A) in side by side replication instead of guanine (Thou) with which the original cytosine would take paired.
As uracil is not present in Dna, adenine base pairs with thymine (T). Therefore C-Thou pair is replaced by T-A in side by side replication cycle. Similarly, hypoxanthine results from adenine deamination.
3. Missing Bases:
Cleavage of Due north-glycosidic bond betwixt purine and sugar causes loss of purine base from Deoxyribonucleic acid. This is called depurination. This apurinic site becomes non-coding lesion.
4. Chemical Modification of Bases:
Chemical modification of any of the four bases of Dna leads to modified bases. Methyl groups are added to various bases. Guanine forms 7- methylguanine, three-methylguanine. Adenine forms 3-methyladenine. Cytosine forms v- Methylcytosine.
Replacement of amino grouping by a keto group converts 5-methylcytosine to thymine.
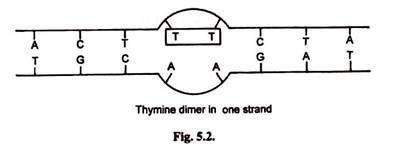
5. Formation of Pyrimidine Dimers (Thymine Dimers):
Formation of thymine dimers is very mutual in which a covalent bond (cyclobutyl ring) is formed between adjacent thymine bases. This leads to loss of base pairing with contrary stand. A bacteria may have thousands of dimers immediately after exposure to ultraviolet radiations.
6. Strand Breaks:
Sometimes phosphodiester bonds break in one strand of DNA helix. This is caused past various chemicals like peroxides, radiations and past enzymes like DNases. This leads to breaks in Deoxyribonucleic acid backbone. Single strand breaks are more common than double strand breaks.
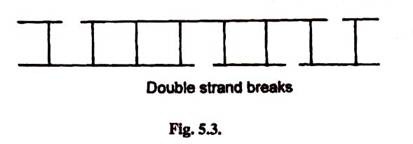
Sometimes X-rays, electronic beams and other radiations may cause phosphodiester bonds breaks in both strands which may not be directly opposite to each other. This leads to double strand breaks.
Some sites on Dna are more susceptible to damage. These are chosen hot-stops.
Repair Mechanisms:
Almost kinds of damage create impediments to replication or transcription. Altered bases cause mispairing and can crusade permanent alteration to DNA sequence after replication.
In club to maintain the integrity of information contained in it, the Deoxyribonucleic acid has various repair mechanisms.
1. Direct Repair:
The harm is reversed by a repair enzyme which is called photoreactivation. This mechanism involves a light dependant enzyme called Deoxyribonucleic acid photolyase. The enzyme is present in almost all cells from leaner to animals. It uses energy from the absorbed lite to cleave the C-C bond of cyclobutyl ring of the thymine dimers. In this way thymine dimers are monomerized.
2. Excision Repair:
It includes base excision repair and nucleotide excision repair. Base excision repair arrangement involves an enzyme called N-glycosylase which recognizes the abnormal base of operations and hydrolyes glycosidic bond between it and sugar.
Some other enzyme, an endonuclease cleaves the DNA backbone on the 5′-side of the aberrant base. And then the Deoxyribonucleic acid polymerase by its exonuclease activity removes the abnormal base. DNA polymerase then replaces it with normal base and Deoxyribonucleic acid ligase seals the region.
Nucleotide repair system includes three steps, incision, excision and synthesis. Incision is done by endonuclease enzyme precisely on either side of the damaged patch of the strand. In this manner damaged portion of the strand is broken.
Endonuclease enzymes involved are UvrA, UvrB which recognize the damaged stretch of the strand. UvrC makes two cuts (incision) on either side. Exonuclease removes the damaged strand. Enzyme involved is UvrD.
Afterwards, DNA polymerase synthesizes the new strand past using complementary strand every bit a template. DNA ligase forms phosphodiester bonds which seal the ends on newly synthesized strand.
iii. Mismatch Base of operations Repair:
Sometimes wrong bases are incorporated during replication process, G-T or C-A pairs are formed. The incorrect base of operations is always incorporated in the daughter strand but. Therefore in social club to distinguish the two strands for the purpose of repair, the adenine bases of the template strand are labelled or tagged past methyl groups. In this way the newly replication Deoxyribonucleic acid helix is hemimethylated. The excision of wrong bases occur in the non-methylated or girl strand.
4. Recombination Repair or Retrieval System:
In thymine dimer or other type of damage, Dna replication cannot proceed properly. A gap opposite to thymine dimer is left in the newly synthesized daughter strand. The gap is repaired by recombination mechanism or retrieval machinery called also sister strand exchange.
During replication of DNA ii identical copies are produced. Replicating Deoxyribonucleic acid molecule has four strands A, B, C and D. Strands A and C accept same Dna , sequence. Strands B and D also have aforementioned sequence as they are identical. A thymine dimer is present in strand A. The replication fork passes the dimmer as it cannot form hydrogen bonds with incoming adenine bases, thus creating a gap in the newly synthesized strand B.
In recombination repair organisation a short identical segment of DNA is retrieved from strand D and is inserted into the gap of strand B. Merely this creates a gap in strand D which is easily filled up by Deoxyribonucleic acid polymerase using normal strand C as a template. This event is dependent on the activeness of a special poly peptide Rec A. The Rec A protein plays its role in retrieving a portion of the complementary strand from other side of the replication fork to make full the gap. Rec A is a strand exchange protein.
After filling both gaps, thymine is monomerised. And then in this repair machinery a portion of DNA strand is retrieved from the normal homologous DNA segment. This is also known every bit daughter strand gap repair mechanism.
5. SOS Repair Mechanism:
Sometimes the replicating machinery is unable to repair the damaged portion and bypasses the damaged site. This is known every bit translesion synthesis as well chosen bypass system and is emergency repair system. This mechanism is catalyzed past a special class of DNA polymerases called Y-family of Deoxyribonucleic acid polymerases which synthesized Dna directly across the damaged portion.
Which Of The Following Images Represents Newly Synthesized Dna That Can Be Repaired,
Source: https://www.biologydiscussion.com/dna/dna-damage-types-and-repair-mechanisms-with-diagram/16332
Posted by: sevignymues1965.blogspot.com


0 Response to "Which Of The Following Images Represents Newly Synthesized Dna That Can Be Repaired"
Post a Comment